Jupyter notebooks batch execution
Date: 05/10/2021
Author: Sébastien Gardoll
Keywords: cluster jupyter notebook batch nbconvert
Basic batch execution of notebooks
With Jupyter, comes a very interesting command: jupyter-nbconvert (see here for another kind of installation). This command allows the conversion of a notebook file (*.ipynb) into a python, pdf, html, latex, rst, etc. script and especially the execution of a notebook (for a list of all formats, type jupyter-nbconvert --help).
The Nbconvert command is very interesting if you want to run a notebook as a batch job on the mesocenter clusters. Knowing that cluster job orchestrators only run scripts, nbconvert is a real deal: it saves you to manage a scripted version of your notebook, just for its execution on clusters (management that inevitably comes with update problems).
The command line is simple by default (for the options, visit this page and visit this page for the templating system). Assuming you have the command in your module environment, the example below runs the notebook named myfile.ipynb and the result is saved in a file named myfile.nbconvert.ipynb. More information at this page.
jupyter-nbconvert --to notebook --ExecutePreprocessor.timeout=-1 --execute myfile.ipynb
All you have to do, is to integrate this command line into a bootstrap Bash script and submit it as a batch job to the cluster scheduler.
Info
The option --ExecutePreprocessor.timeout=-1 disables the cell execution timeout whose default value is 30 seconds.
Info
If you want the result to be saved directly in the notebook, add the --clear-output option. But it is strongly recommended to use the --inplace option instead to avoid mixing the result with the previous one.
Conditional execution of notebook cells
Conditional execution of notebook cells is possible using the nbconvert command (see above for its introduction). The first step is to mark the cells that you want to ignore during the execution of the notebook, using the Jupyter marking system:
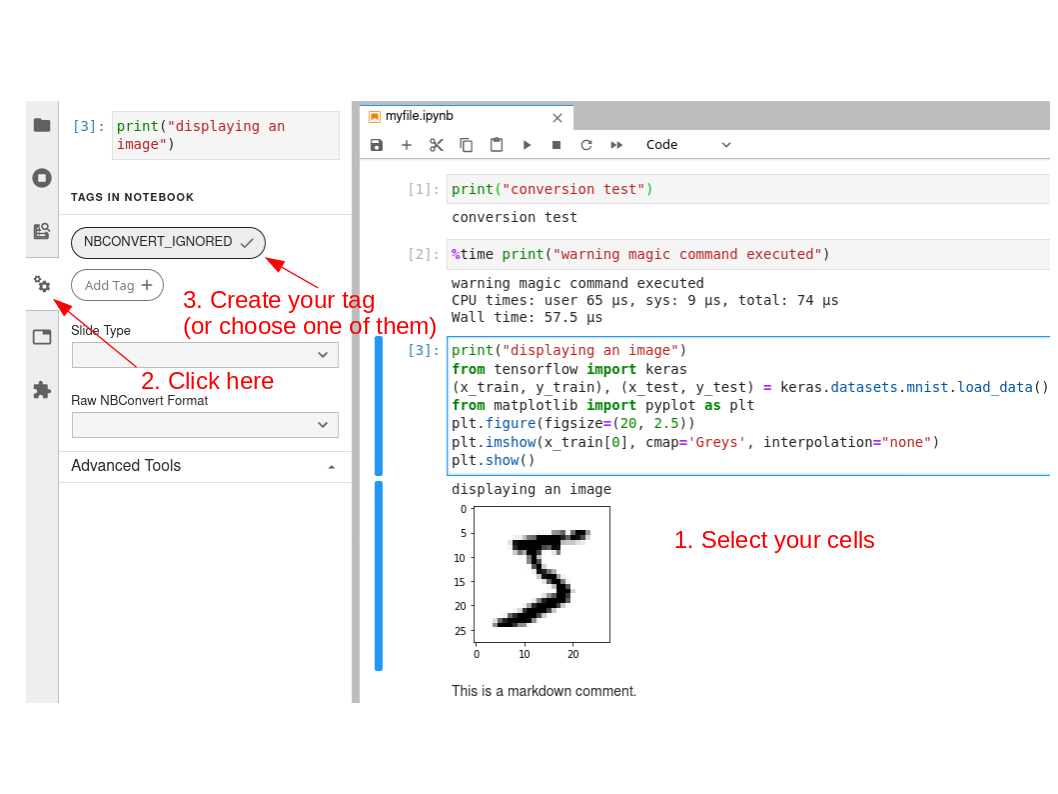
Save the notebook, then run the notebook with the following command line:
jupyter-nbconvert --to notebook --ExecutePreprocessor.timeout=-1 --TagRemovePreprocessor.enabled=True --TagRemovePreprocessor.remove_cell_tags 'NBCONVERT_IGNORED' --execute myfile.ipynb
Warning
Never use the --inplace and --clear-output options unless you want to lose the marked cells forever!!!
Info
The NBCONVERT_IGNORED marker does not matter but it must be consistent with the command line.
Info
Nbconvert has a lot of options. Combined with the Jupyter marking system, it is possible to emulate conditional code generation (a bit like the C language preprocessor).